Each year more than 500,000 infants in the U.S.A. are born prematurely, up to a third of whom will develop a neurodevelopmental disorder (NDD). NDDs range from autism to cerebral palsy and are caused by alterations in brain connectivity. Chronic hypoxic exposure, which is a near-ubiquitous complication of prematurity, is one of the primary risks for NDDs. Hypoxia is a large reduction of oxygen. The hypoxia occurs during a developmental epoch (period of time) characterized by widespread axon pathfinding and synaptogenesis (which broadly refer to as connectivity development) in the CNS. Despite its importance, the molecular mechanisms by which hypoxia disrupts CNS connectivity development are poorly understood.
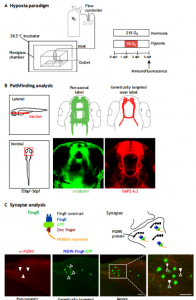
Dr. Son and his colleagues have developed a novel zebrafish model of chronic hypoxia to study effects of prematurity. To induce hypoxia, embryonic zebrafish are placed in a sealed Plexiglass chamber connected via a controller that monitors and adjusts nitrogen gas flow to a desired oxygen pressure (pO2) set point (Biospherix, Inc.) (Fig. 1A). They showed that hypoxic injury specifically disrupts axon pathfinding, which is a quantifiable defect that occurs during the period of axon extension from 24-36 hours post-fertilization (hpf) (Figure 1B). Their hypoxia model disrupts CNS connectivity development, but they have shown that it does not cause an increase in apoptosis or grossly impact other aspects of neurogenesis or fate determination, which is similar to the effects of prematurity in human infants, and to the mouse models of chronic hypoxia. Additionally, to monitor and visualize synapses, Dr. Son and his colleagues have modified and implemented the use of fibronectin intrabodies generated by mRNA display (FingRs) in zebrafish (Fig. 1C). Dr. Son and his colleagues were the first to make transgenic FingR animals; and the first to demonstrate FingR use in live, behaving animals. FingRs are an in vivo, the targetable genetic method for monitoring and visualizing synapses. FingRs were initially tested in cell culture and transiently in hippocampal slices, targeting synaptic proteins such as excitatory postsynaptic density 95 (PSD-95) or inhibitory Gephyrin synaptic protein (GPHN). Previously no reliable genetic technique was available to label postsynaptic targets in live animals. With FingRs they can monitor synapses in genetically defined groups of neurons as opposed to labeling all of the synapses when using pan-labeling immunohistochemistry; they can track changes in live animals; FingRs can be visualized post-fixation as well. With FingRs, they have performed extensive controls regarding the specificity of their expression and localization. They have confirmed FingR postsynaptic localization to synapses; and shown that their expression does not impair expression of other synaptic proteins or behavior, which is consistent with the initial published work on FingRs as well as subsequent use.
The small vertebrate zebrafish (Danio rerio) has unique advantages for this project. Experimentally, zebrafish has straightforward gene function manipulation for transgenesis and CRISPR knockdown, and facile imaging, similar to invertebrates. Dr. Son and his colleagues have generated multiple transgenic lines that they actively use for research experiments that would not financially or genetically be feasible in other vertebrate models. Zebrafish have been used as models for neurodevelopmental disorders ranging from epilepsy to hydrocephalus and recently have been used for discovery of treatments for human neurological diseases as well.
To read more about Dr. Son’s research, please keep on the lookout for the release of this article “Hypoxia and connectivity in the developing vertebrate nervous system” in the journal Disease Models & Mechanisms.